Paper 04: rROC theoretic with FPR
paper_04_rROC_theoretic_FPR.Rmd
library(restrictedROC)
library(ggplot2)
size_factor <- .34
main_height <- 20
main_width <- 20
dir.create("res/paper", recursive = TRUE)
#> Warning in dir.create("res/paper", recursive = TRUE): 'res/paper' already
#> exists
main_plotname <- "res/paper/rROC_fpr"Do not use the code!
This code here is not intended to be used but rather to show the resulting plots. The usage is somewhat different to what I would suggest. Look at the pictures, not at the code.
plots_including_fpr <- function(rroc_theo_result, rroc_ylim = NA) {
plot_x_fpr <- ggplot(
rroc_theo_result[["rroc"]][["single_rROC"]][["performances"]],
aes(x = threshold, y = fpr_global)
) +
geom_point() +
ylab("False positive rate") +
xlab("x threshold") +
ggpubr::theme_pubr()
srroc <- rroc_theo_result[["rroc"]][["single_rROC"]]
max_th <- srroc$max_total$threshold
max_rzauc <- srroc$max_total$rzAUC
max_fpr <- srroc$performances[srroc$performances$threshold == max_th, ][["fpr_global"]]
max_tpr <- srroc$performances[srroc$performances$threshold == max_th, ][["tpr_global"]]
if (!all(is.na(rroc_ylim))) {
tmp <- rroc_theo_result[["rroc"]][["plots"]] + ggplot2::ylim(rroc_ylim)
} else {
tmp <- rroc_theo_result[["rroc"]][["plots"]]
}
return(list(
tmp,
tmp + ggplot2::theme(legend.position = "none"),
ggpubr::as_ggplot(ggpubr::get_legend(tmp)),
plot_x_fpr,
patchwork::wrap_plots(list(
"A" = tmp[[1]],
"B" = tmp[[3]] +
geom_segment(x = max_fpr, xend = max_fpr, y = -Inf, yend = max_tpr, col = "red"),
"C" = plot_x_fpr +
geom_segment(x = max_th, xend = max_th, y = -Inf, yend = max_fpr, col = "red") +
geom_segment(x = -Inf, xend = max_th, y = max_fpr, yend = max_fpr, col = "red"),
"D" = tmp[[4]] +
geom_segment(x = max_fpr, xend = max_fpr, y = -Inf, yend = max_rzauc, col = "red") +
ggplot2::theme(legend.position = "none")
)) +
patchwork::plot_layout(design = "AB\nCD")
))
}
n_positives <- 100
n_negatives <- 100
# 1. Random
filename <- file.path(paste0(main_plotname, "_random.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) qnorm(x, mean = 5, sd = 1),
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 1),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo, rroc_ylim = c(-1, 1)))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""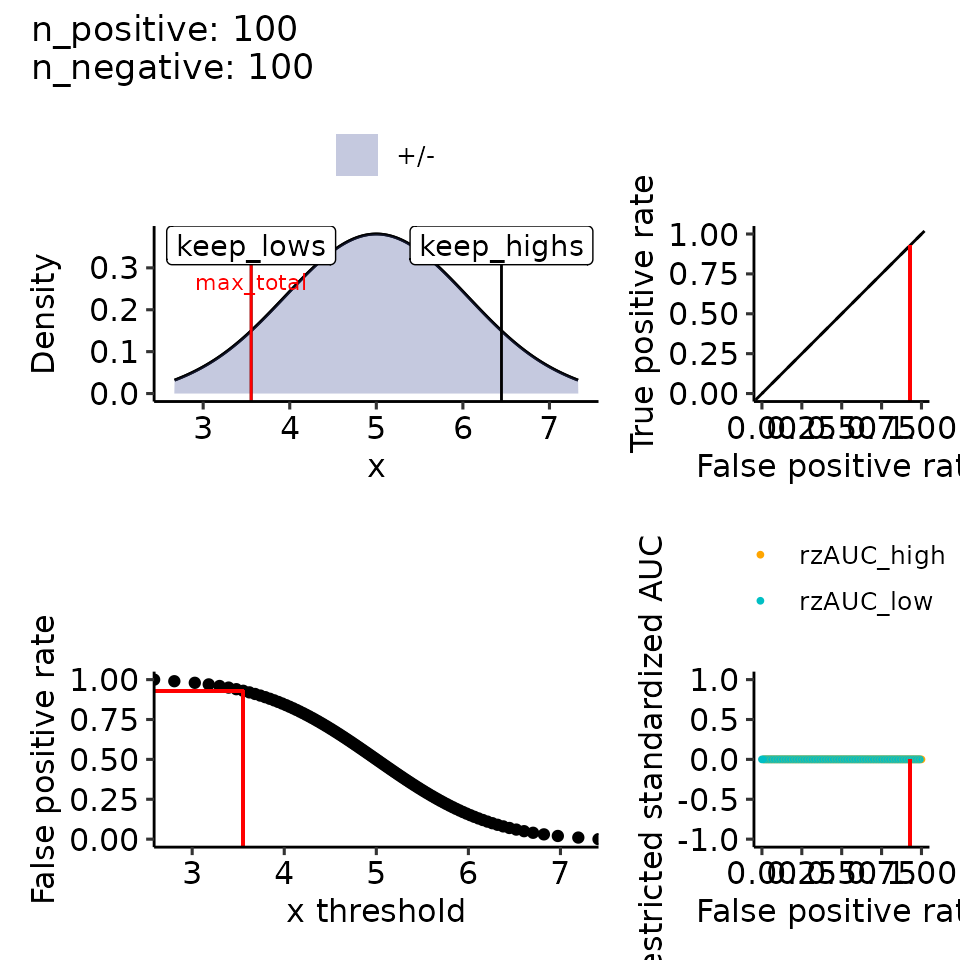
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""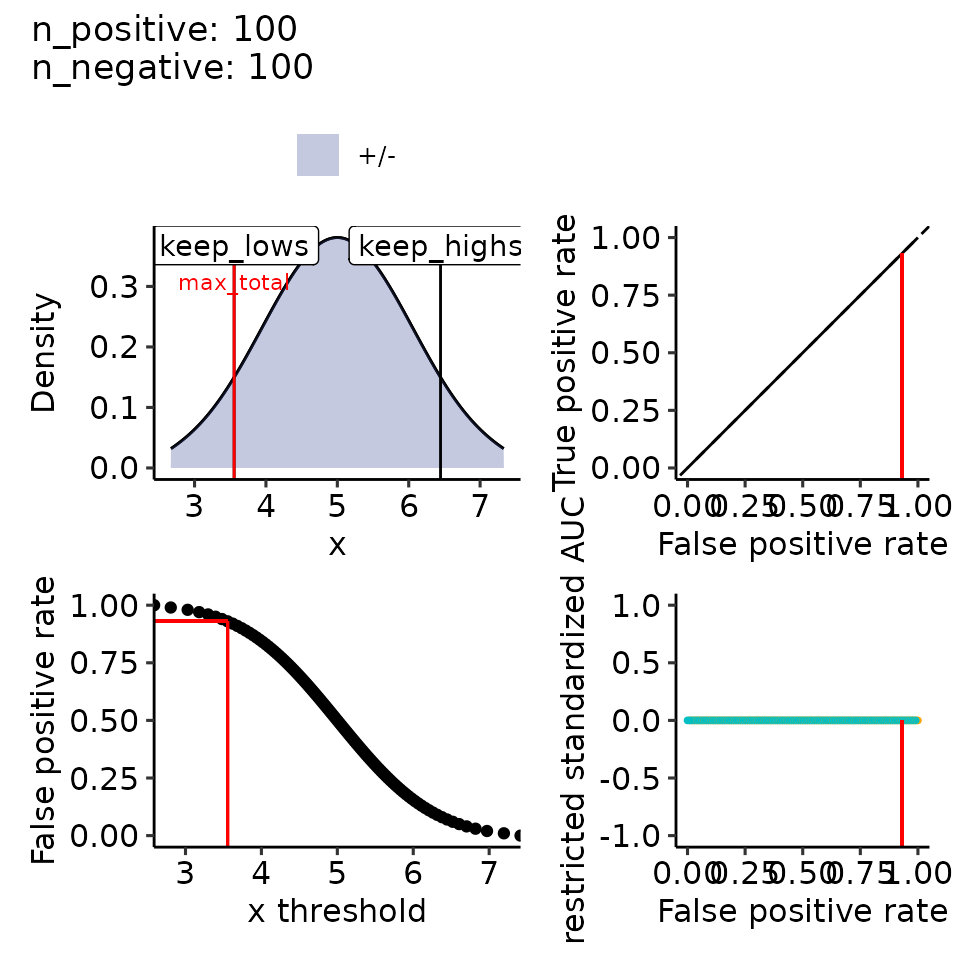
#>
#> [[3]]
#>
#> [[4]]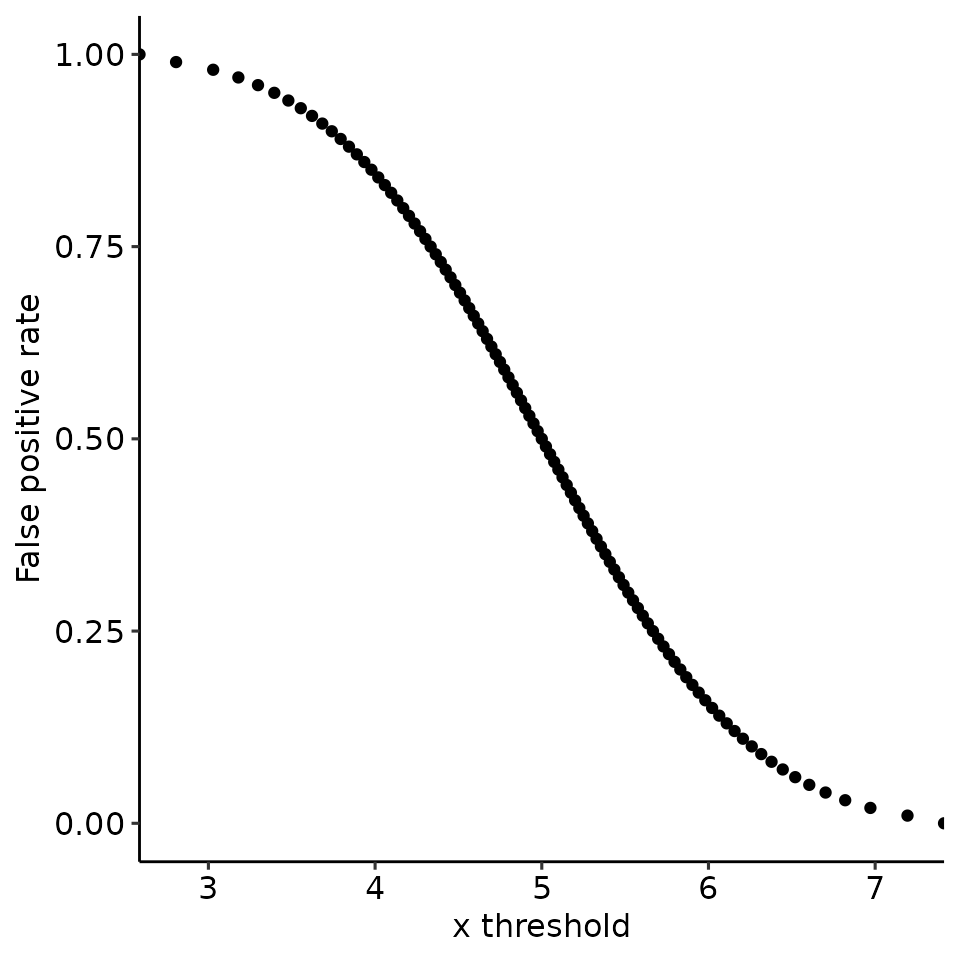
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""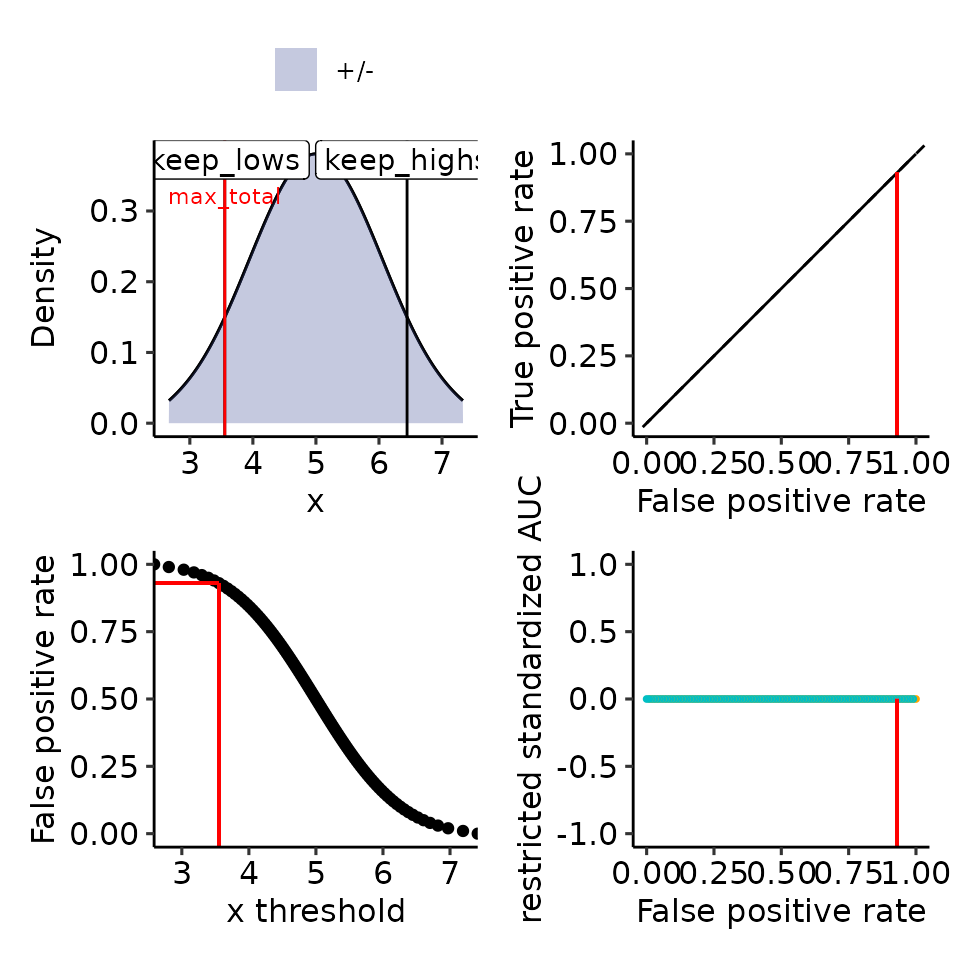
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#> $data$negative
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""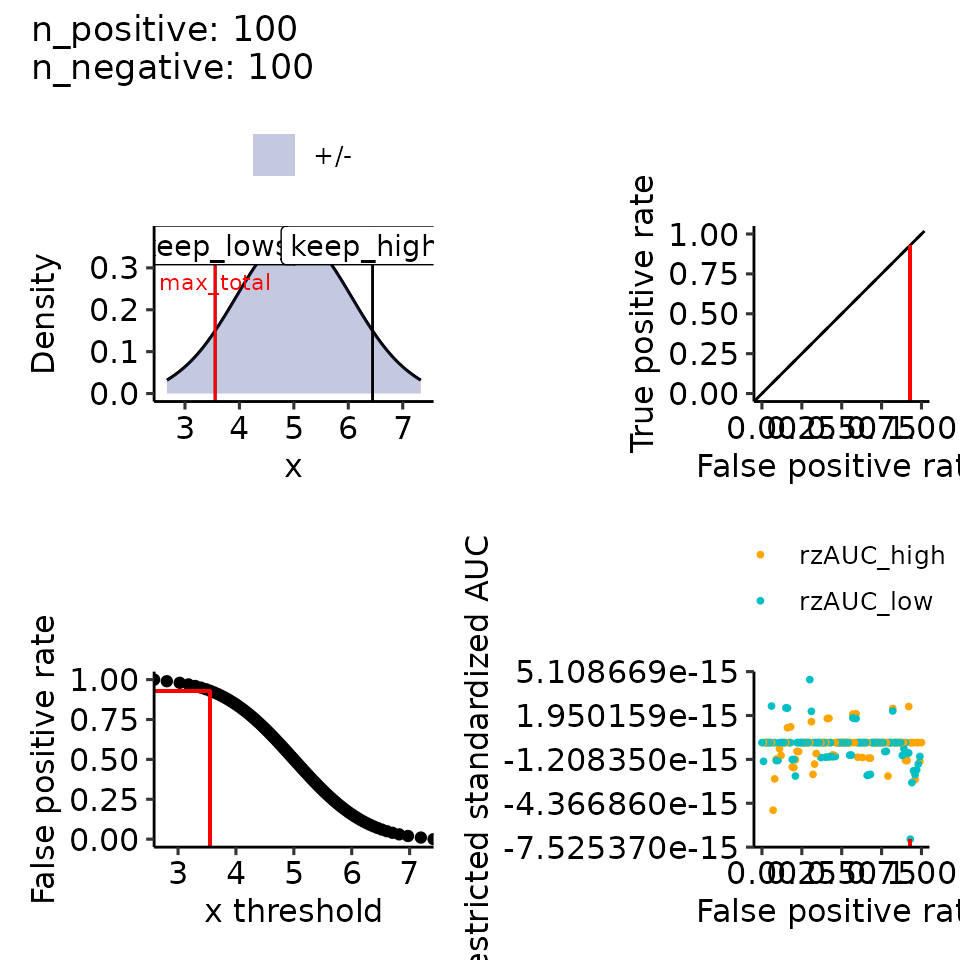
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 101 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.5 100 100 1 0.00168
#> 2 2.81 0.5 99 99 1.02 0.00169
#> 3 3.03 0.5 98 98 1.04 0.00171
#> 4 3.18 0.5 97 97 1.06 0.00173
#> 5 3.30 0.5 96 96 1.09 0.00175
#> 6 3.40 0.5 95 95 1.11 0.00176
#> 7 3.48 0.5 94 94 1.13 0.00178
#> 8 3.55 0.5 93 93 1.16 0.00180
#> 9 3.62 0.5 92 92 1.18 0.00182
#> 10 3.68 0.5 91 91 1.21 0.00184
#> # ℹ 91 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.5 0.5 0 1
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.5 0.0255102 -4.865767e-15 1 6.445696
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.5 0.0255102 -6.951096e-15 1 3.554304
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.5 0.0255102 -6.951096e-15 1 3.554304 low
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.5
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()
filename <- file.path(paste0(main_plotname, "_posGTneg.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) qnorm(x, mean = 6, sd = 1),
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 1),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""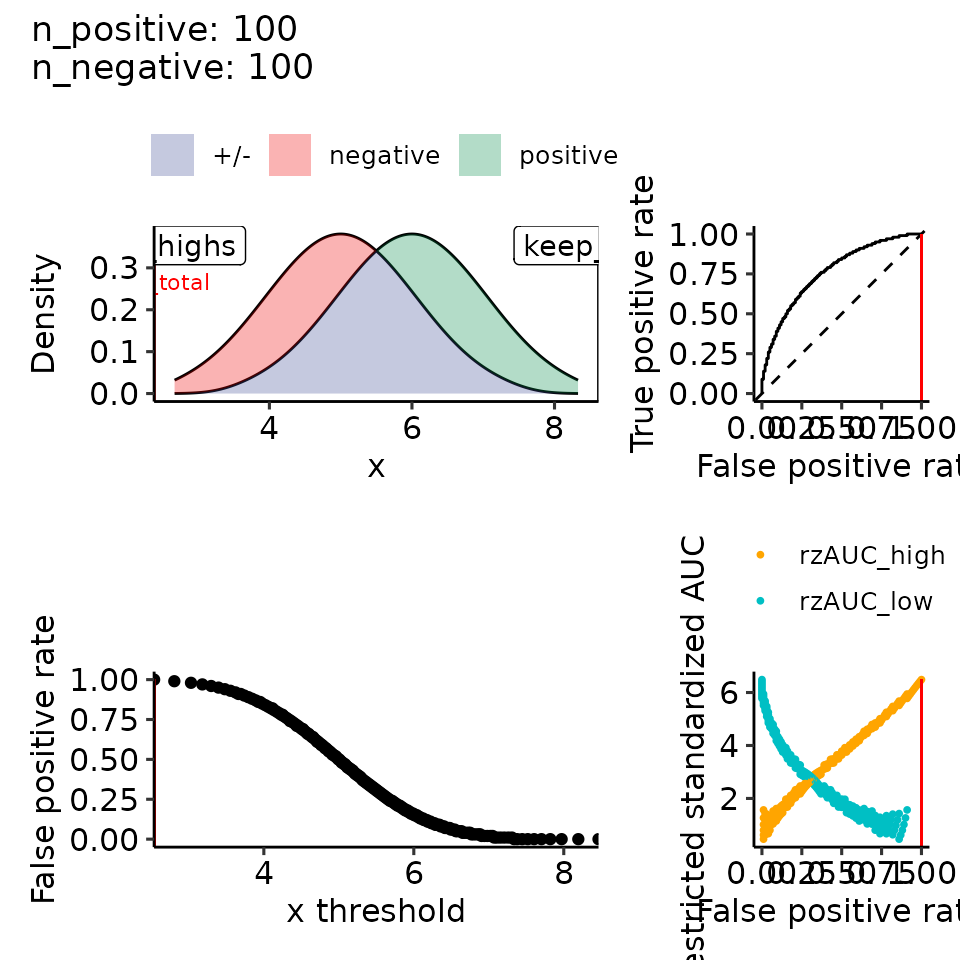
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""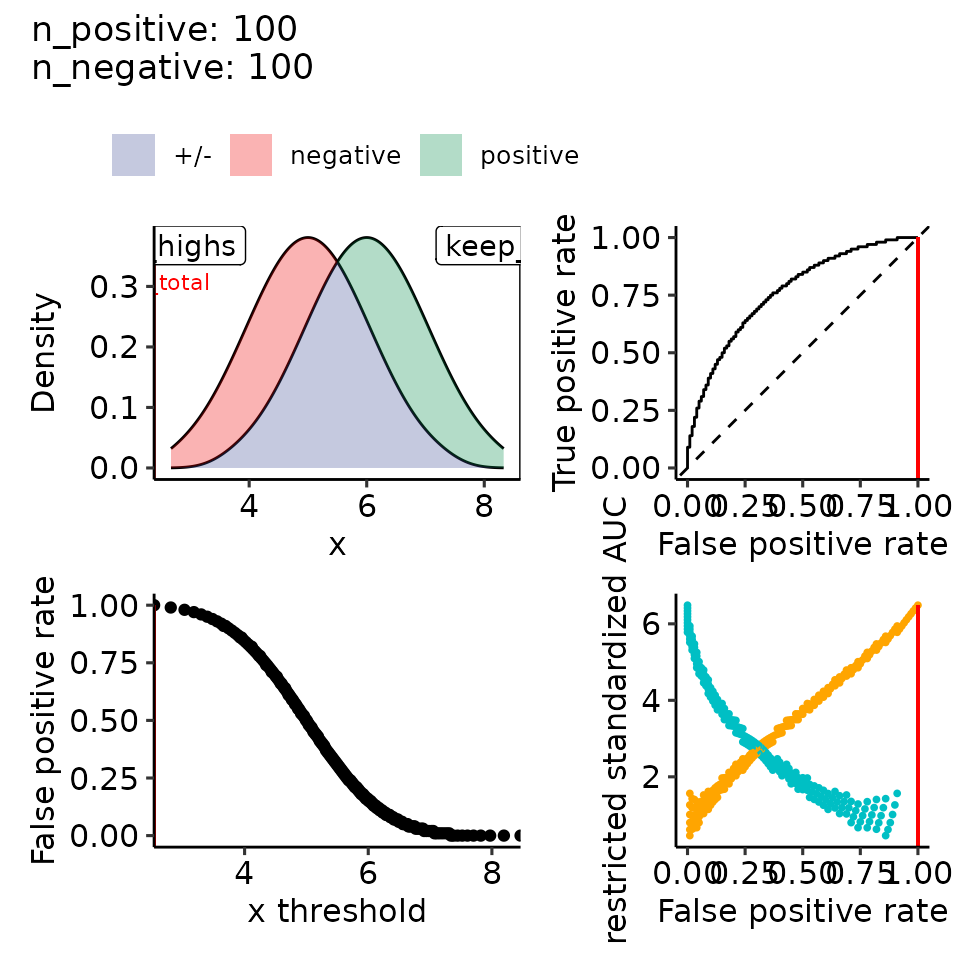
#>
#> [[3]]
#>
#> [[4]]
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""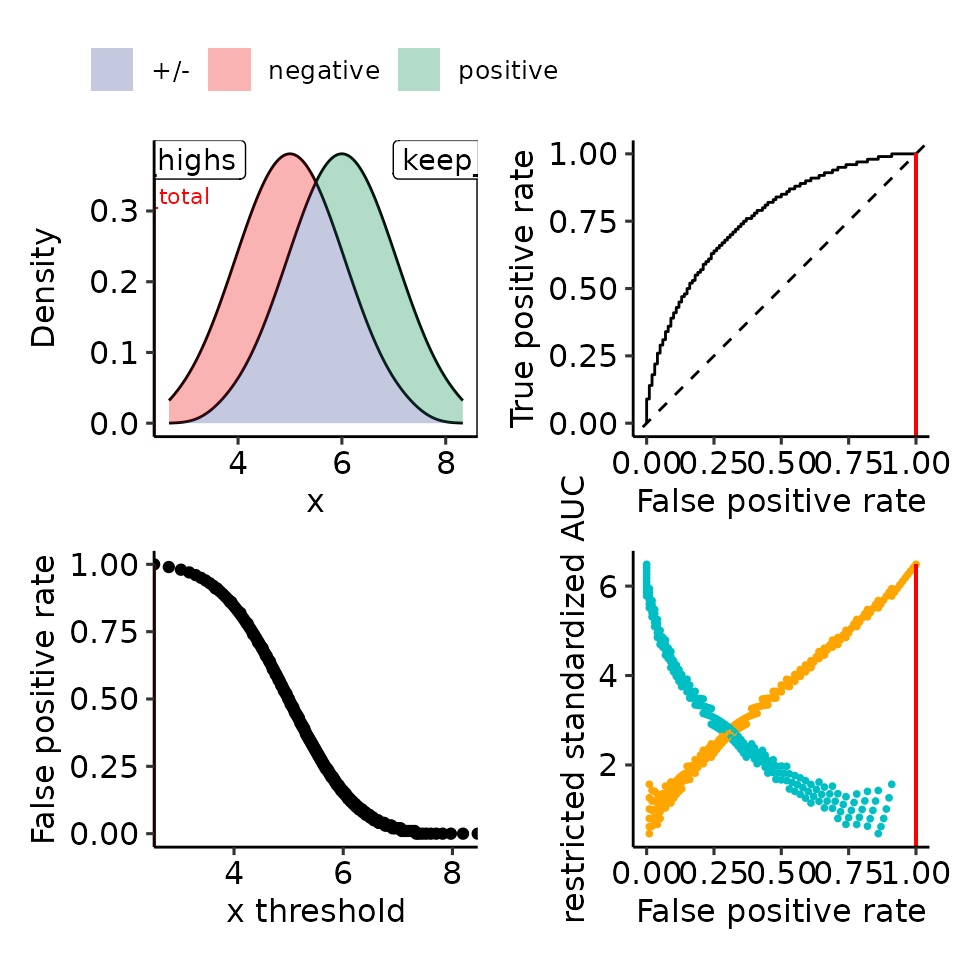
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 3.669921 3.942144 4.114823 4.244699 4.350327 4.440220 4.519027 4.589580
#> [9] 4.653737 4.712786 4.767659 4.819053 4.867503 4.913432 4.957176 4.999010
#> [17] 5.039162 5.077822 5.115150 5.151284 5.186343 5.220429 5.253633 5.286033
#> [25] 5.317700 5.348698 5.379082 5.408903 5.438208 5.467037 5.495431 5.523423
#> [33] 5.551047 5.578332 5.605307 5.631997 5.658428 5.684623 5.710603 5.736388
#> [41] 5.762000 5.787457 5.812776 5.837976 5.863074 5.888085 5.913027 5.937915
#> [49] 5.962764 5.987591 6.012409 6.037236 6.062085 6.086973 6.111915 6.136926
#> [57] 6.162024 6.187224 6.212543 6.238000 6.263612 6.289397 6.315377 6.341572
#> [65] 6.368003 6.394693 6.421668 6.448953 6.476577 6.504569 6.532963 6.561792
#> [73] 6.591097 6.620918 6.651302 6.682300 6.713967 6.746367 6.779571 6.813657
#> [81] 6.848716 6.884850 6.922178 6.960838 7.000990 7.042824 7.086568 7.132497
#> [89] 7.180947 7.232341 7.287214 7.346263 7.410420 7.480973 7.559780 7.649673
#> [97] 7.755301 7.885177 8.057856 8.330079
#>
#> $data$negative
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""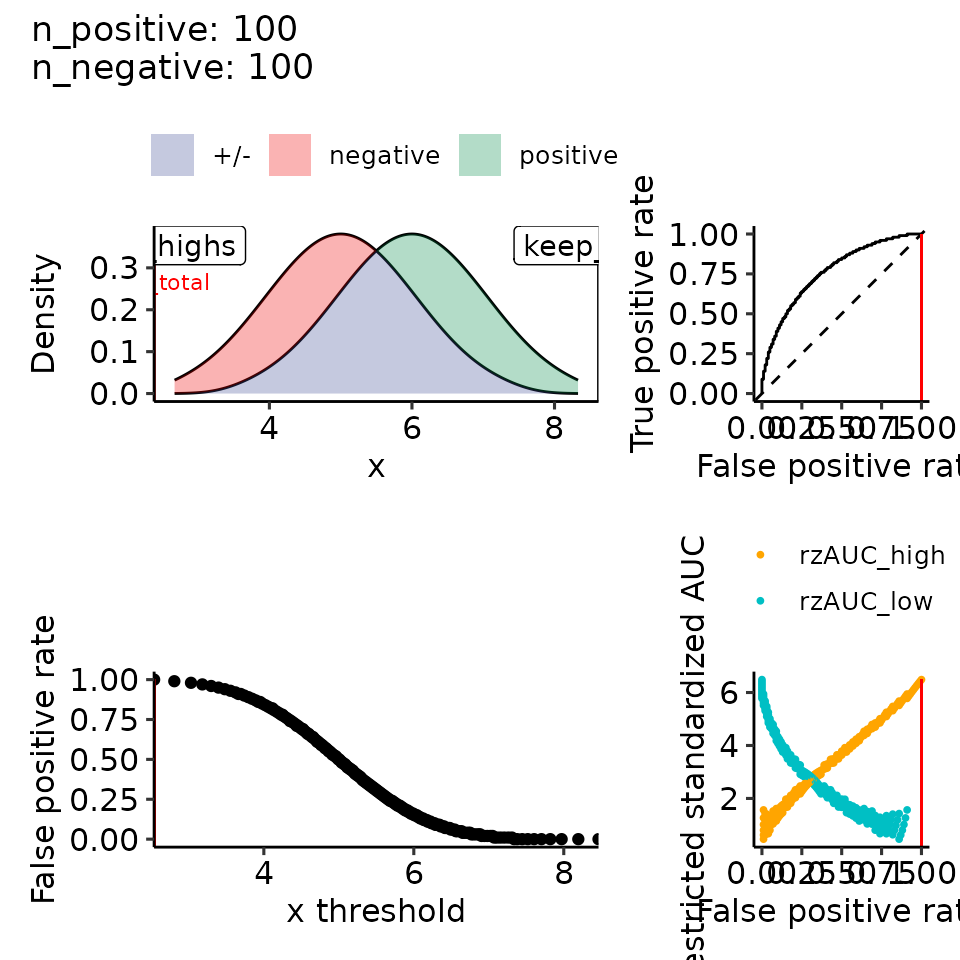
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 201 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.766 100 100 1 0.00168
#> 2 2.81 0.763 100 99 1.01 0.00168
#> 3 3.03 0.761 100 98 1.02 0.00169
#> 4 3.18 0.758 100 97 1.03 0.00170
#> 5 3.30 0.756 100 96 1.04 0.00171
#> 6 3.40 0.753 100 95 1.05 0.00172
#> 7 3.48 0.751 100 94 1.06 0.00173
#> 8 3.55 0.748 100 93 1.08 0.00174
#> 9 3.62 0.745 100 92 1.09 0.00175
#> 10 3.66 0.742 100 91 1.10 0.00176
#> # ℹ 191 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.7655 0.7655 6.487197 8.74476e-11
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.7655 0.001675 6.487197 8.74476e-11 -Inf
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.7655 0.001675 6.487197 8.74476e-11 Inf
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.7655 0.001675 6.487197 8.74476e-11 -Inf global
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.7655
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()
filename <- file.path(paste0(main_plotname, "_pos2norm_highdiff.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) {
retvec <- numeric(length(x))
for (i in seq_along(x)) {
if (i %% 4 == 0) {
retvec[i] <- qnorm(x[i], 9, 1)
} else {
retvec[i] <- qnorm(x[i], 5, 1)
}
}
return(retvec)
},
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 1),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""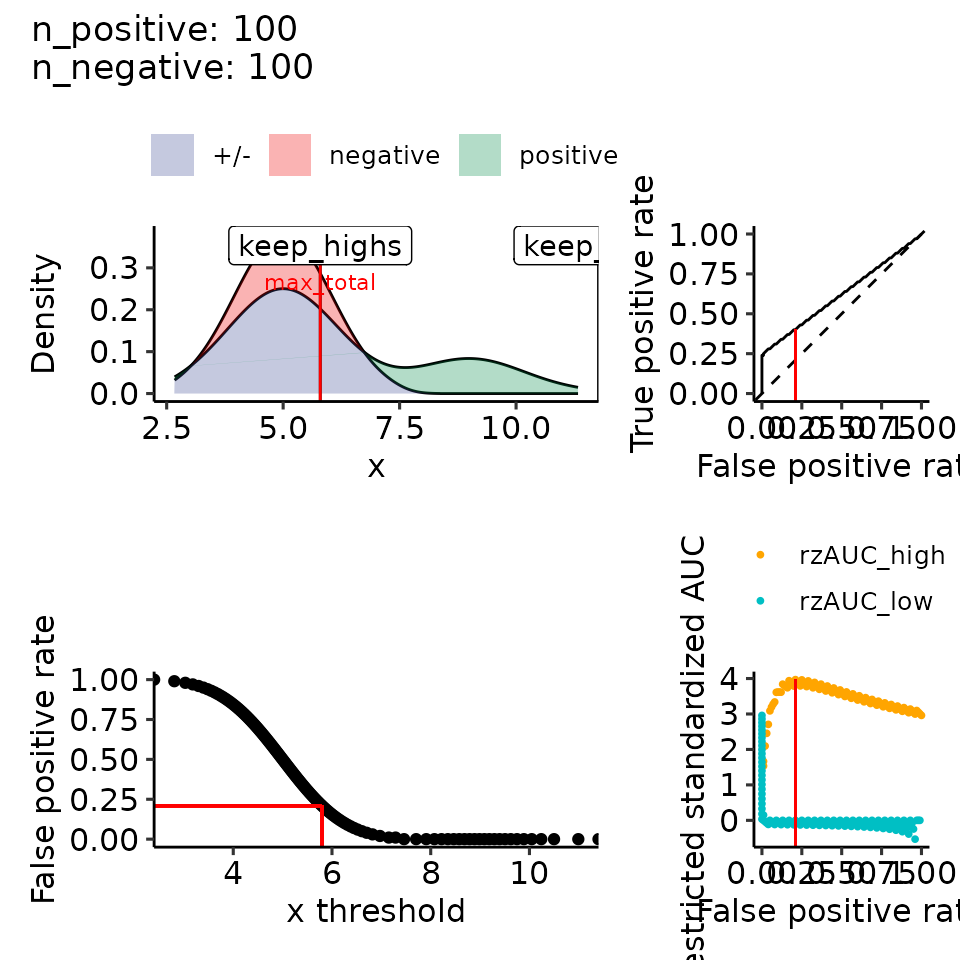
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""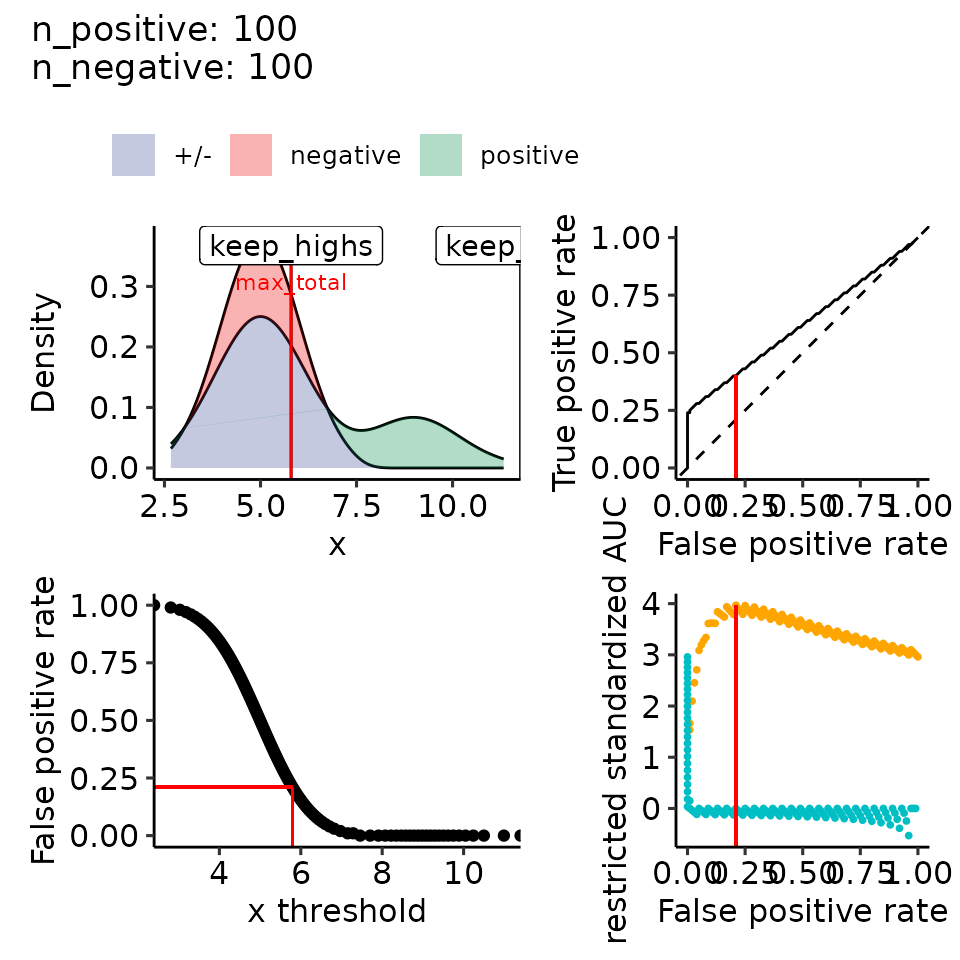
#>
#> [[3]]
#>
#> [[4]]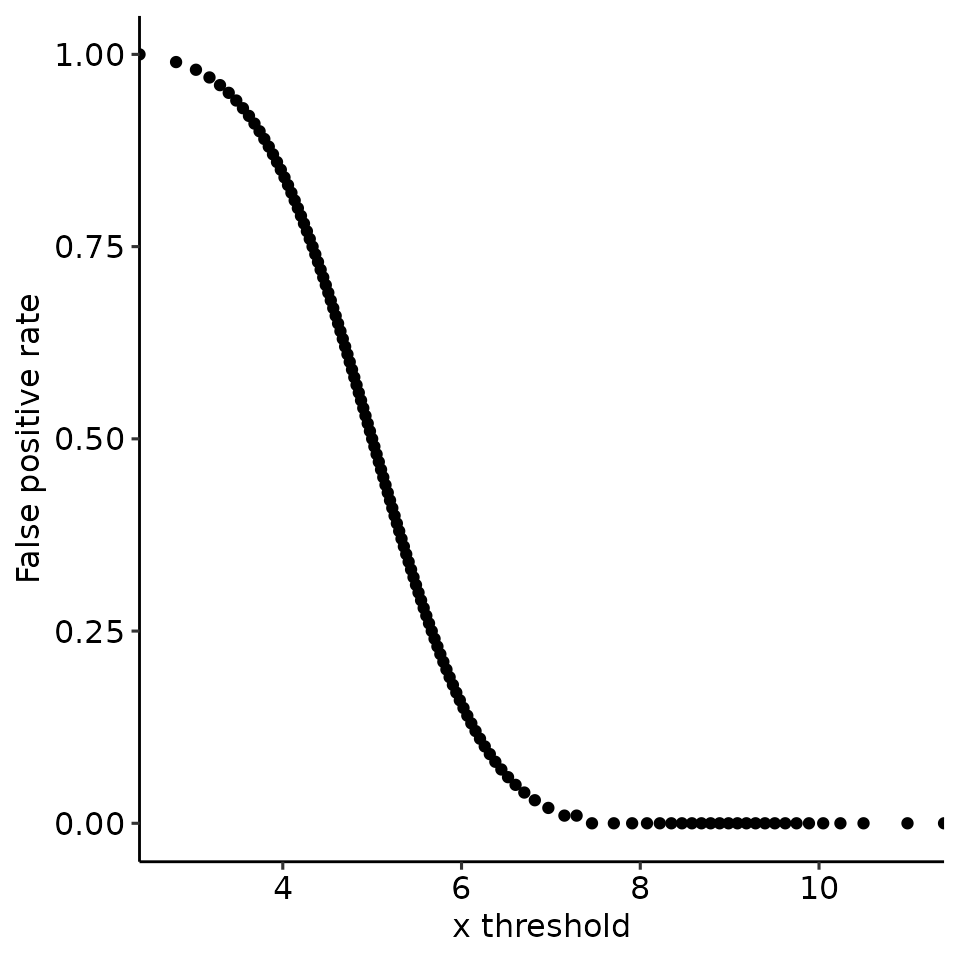
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""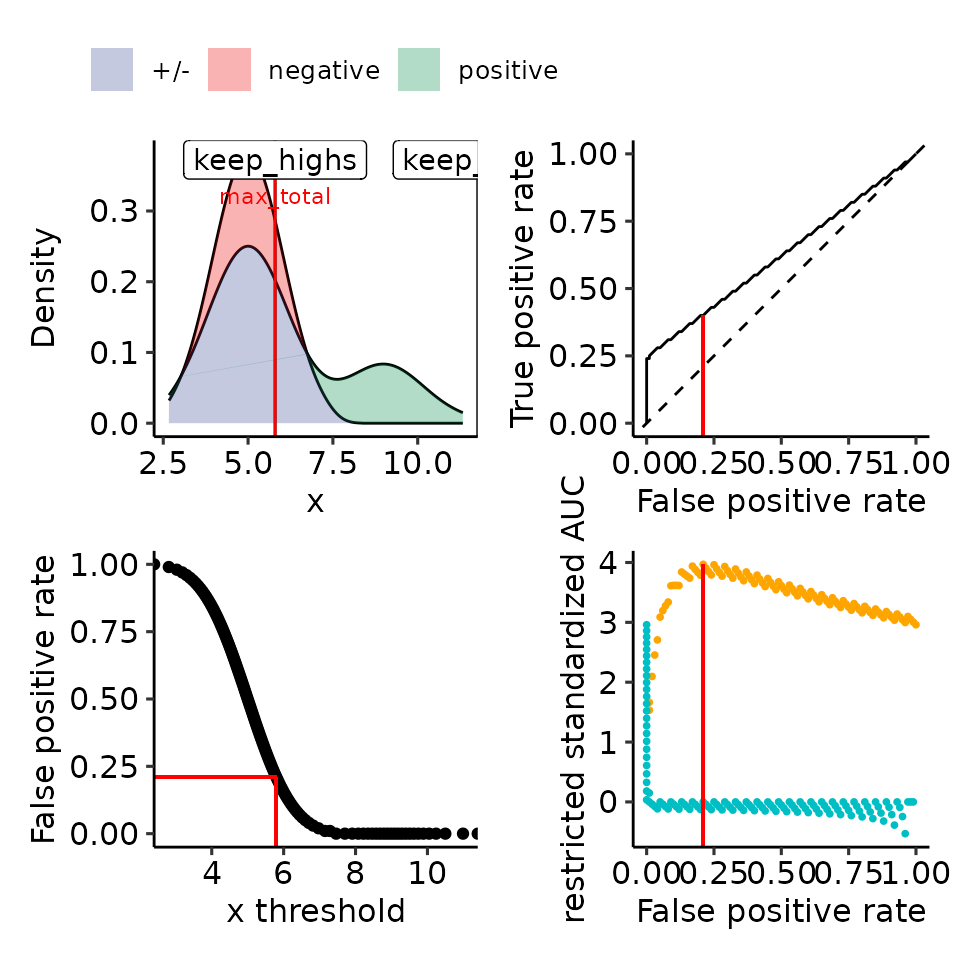
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 2.669921 2.942144 3.114823 7.244699 3.350327 3.440220 3.519027
#> [8] 7.589580 3.653737 3.712786 3.767659 7.819053 3.867503 3.913432
#> [15] 3.957176 7.999010 4.039162 4.077822 4.115150 8.151284 4.186343
#> [22] 4.220429 4.253633 8.286033 4.317700 4.348698 4.379082 8.408903
#> [29] 4.438208 4.467037 4.495431 8.523423 4.551047 4.578332 4.605307
#> [36] 8.631997 4.658428 4.684623 4.710603 8.736388 4.762000 4.787457
#> [43] 4.812776 8.837976 4.863074 4.888085 4.913027 8.937915 4.962764
#> [50] 4.987591 5.012409 9.037236 5.062085 5.086973 5.111915 9.136926
#> [57] 5.162024 5.187224 5.212543 9.238000 5.263612 5.289397 5.315377
#> [64] 9.341572 5.368003 5.394693 5.421668 9.448953 5.476577 5.504569
#> [71] 5.532963 9.561792 5.591097 5.620918 5.651302 9.682300 5.713967
#> [78] 5.746367 5.779571 9.813657 5.848716 5.884850 5.922178 9.960838
#> [85] 6.000990 6.042824 6.086568 10.132497 6.180947 6.232341 6.287214
#> [92] 10.346263 6.410420 6.480973 6.559780 10.649673 6.755301 6.885177
#> [99] 7.057856 11.330079
#>
#> $data$negative
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""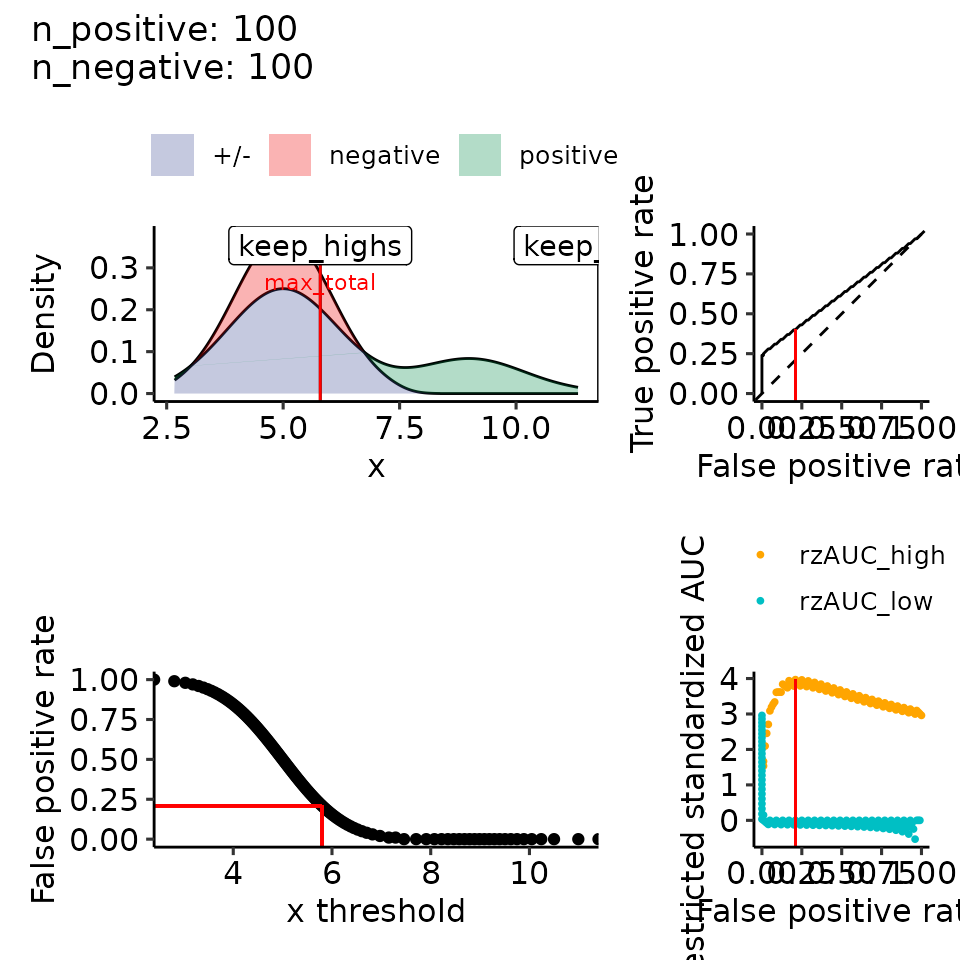
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 126 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.621 100 100 1 0.00168
#> 2 2.81 0.624 99 99 1.02 0.00169
#> 3 3.03 0.626 98 98 1.04 0.00171
#> 4 3.18 0.629 97 97 1.06 0.00173
#> 5 3.30 0.625 97 96 1.07 0.00174
#> 6 3.40 0.627 96 95 1.10 0.00175
#> 7 3.48 0.630 95 94 1.12 0.00177
#> 8 3.55 0.633 94 93 1.14 0.00179
#> 9 3.62 0.629 94 92 1.16 0.00180
#> 10 3.68 0.632 93 91 1.18 0.00182
#> # ℹ 116 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.62115 0.62115 2.960166 0.003074737
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.8113095 0.006150794 3.969418 7.204841e-05 5.796614
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.62115 0.001675 2.960166 0.003074737 Inf
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.8113095 0.006150794 3.969418 7.204841e-05 5.796614 high
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.6212
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()
filename <- file.path(paste0(main_plotname, "_pos2norm_highdiff_v2.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) {
retvec <- numeric(length(x))
for (i in seq_along(x)) {
if (i %% 10 == 0) {
retvec[i] <- qnorm(x[i], 9, 1)
} else {
retvec[i] <- qnorm(x[i], 5, 1)
}
}
return(retvec)
},
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 1),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""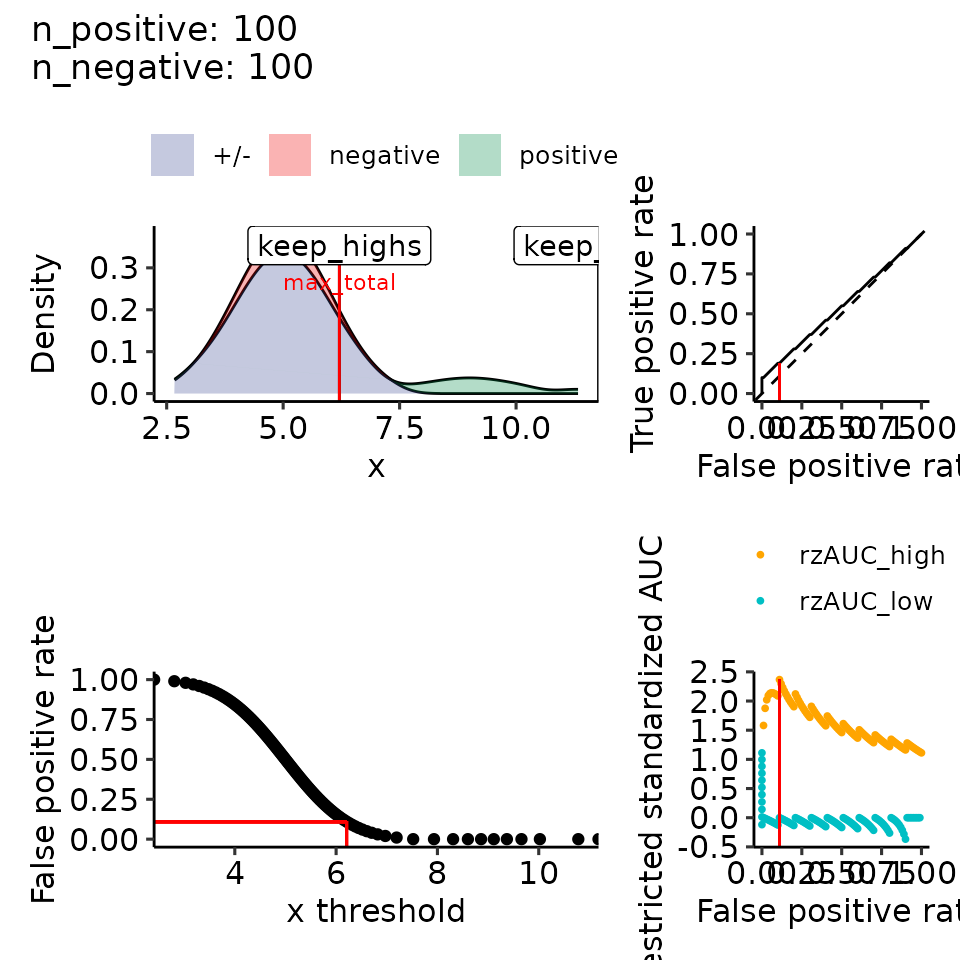
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""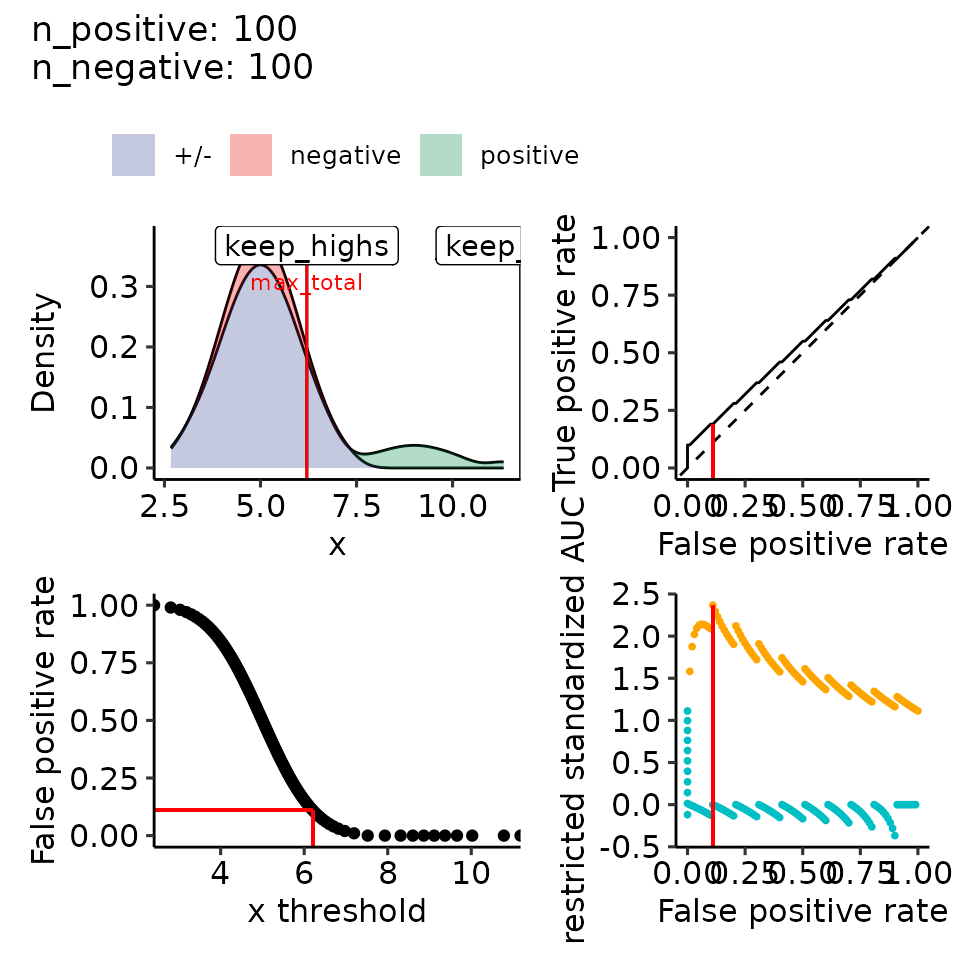
#>
#> [[3]]
#>
#> [[4]]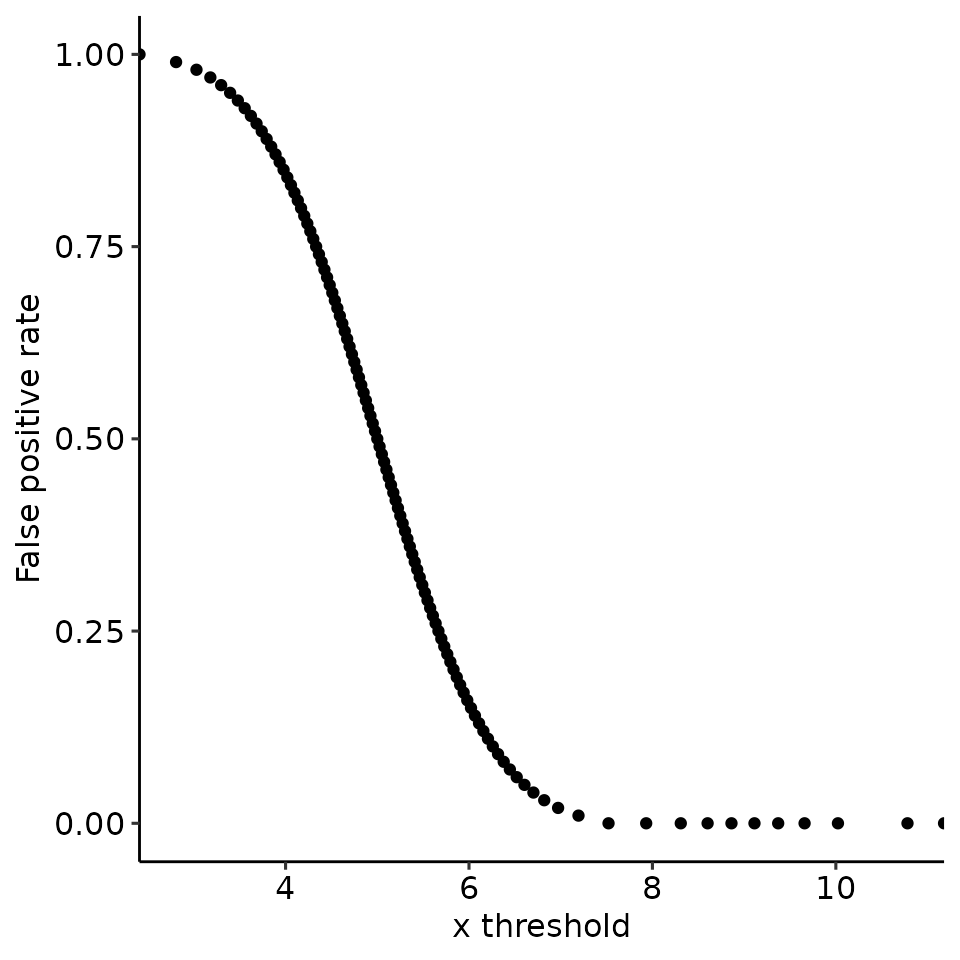
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""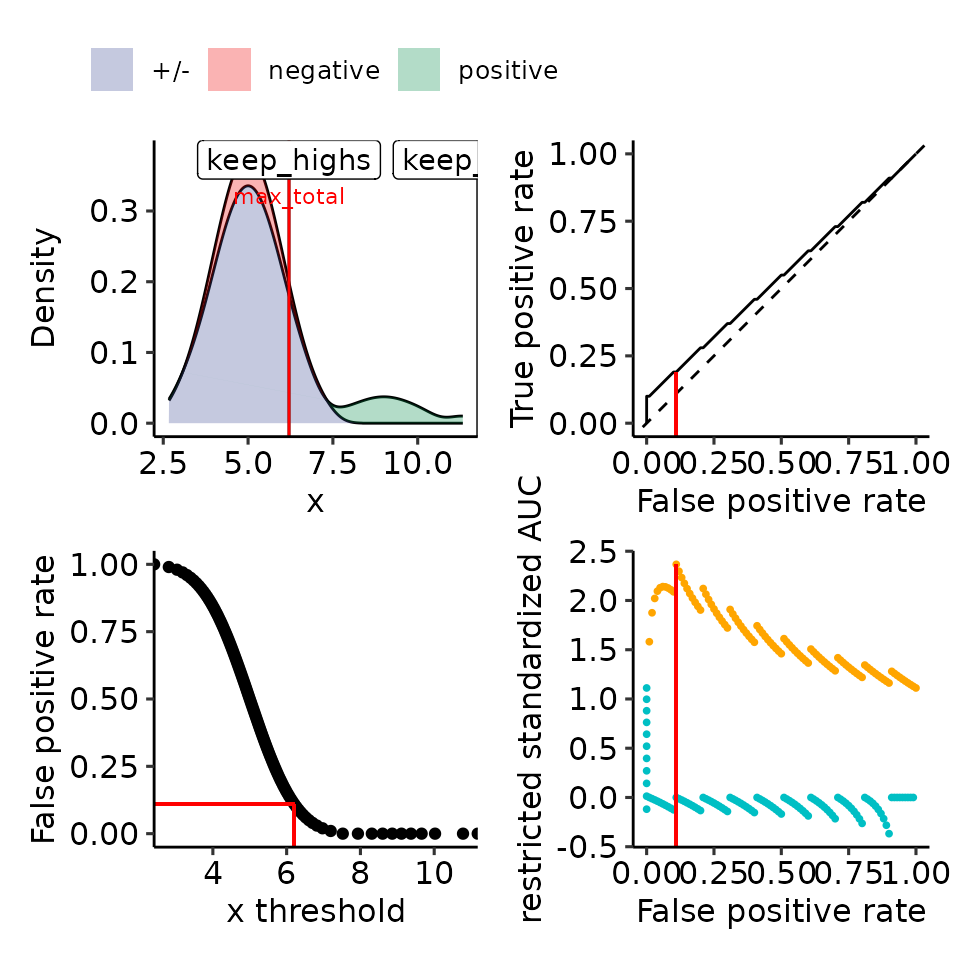
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027
#> [8] 3.589580 3.653737 7.712786 3.767659 3.819053 3.867503 3.913432
#> [15] 3.957176 3.999010 4.039162 4.077822 4.115150 8.151284 4.186343
#> [22] 4.220429 4.253633 4.286033 4.317700 4.348698 4.379082 4.408903
#> [29] 4.438208 8.467037 4.495431 4.523423 4.551047 4.578332 4.605307
#> [36] 4.631997 4.658428 4.684623 4.710603 8.736388 4.762000 4.787457
#> [43] 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915 4.962764
#> [50] 8.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 9.238000 5.263612 5.289397 5.315377
#> [64] 5.341572 5.368003 5.394693 5.421668 5.448953 5.476577 9.504569
#> [71] 5.532963 5.561792 5.591097 5.620918 5.651302 5.682300 5.713967
#> [78] 5.746367 5.779571 9.813657 5.848716 5.884850 5.922178 5.960838
#> [85] 6.000990 6.042824 6.086568 6.132497 6.180947 10.232341 6.287214
#> [92] 6.346263 6.410420 6.480973 6.559780 6.649673 6.755301 6.885177
#> [99] 7.057856 11.330079
#>
#> $data$negative
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""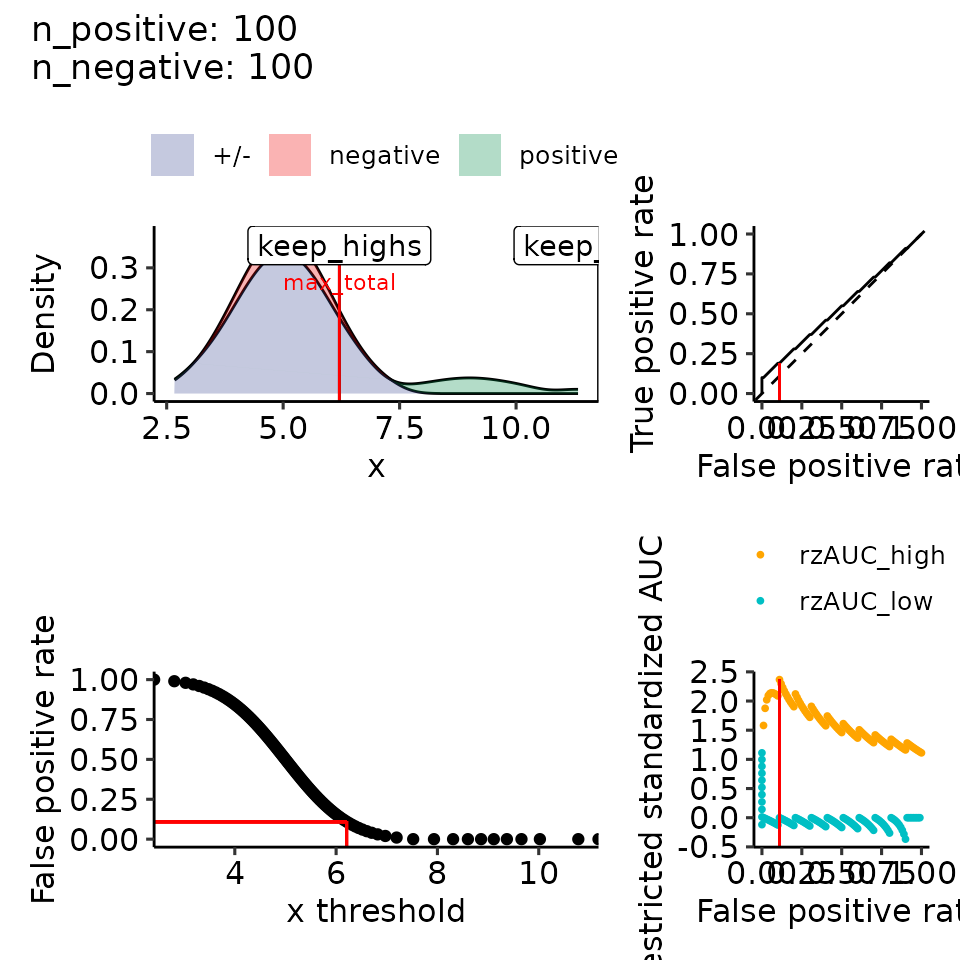
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 111 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.546 100 100 1 0.00168
#> 2 2.81 0.546 99 99 1.02 0.00169
#> 3 3.03 0.547 98 98 1.04 0.00171
#> 4 3.18 0.548 97 97 1.06 0.00173
#> 5 3.30 0.549 96 96 1.09 0.00175
#> 6 3.40 0.550 95 95 1.11 0.00176
#> 7 3.48 0.551 94 94 1.13 0.00178
#> 8 3.55 0.553 93 93 1.16 0.00180
#> 9 3.62 0.554 92 92 1.18 0.00182
#> 10 3.68 0.555 91 91 1.21 0.00184
#> # ℹ 101 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.5455 0.5455 1.111742 0.2662491
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.7631579 0.01236045 2.367006 0.01793265 6.206644
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.5455 0.001675 1.111742 0.2662491 Inf
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.7631579 0.01236045 2.367006 0.01793265 6.206644 high
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.5455
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()
# 4. Different mean + variance
# 4.1 mean: positive > negative, var: positive > negative --> left-skewed
filename <- file.path(paste0(main_plotname, "_posGTneg_posVARGTneg.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) qnorm(x, mean = 6, sd = 2),
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 1),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""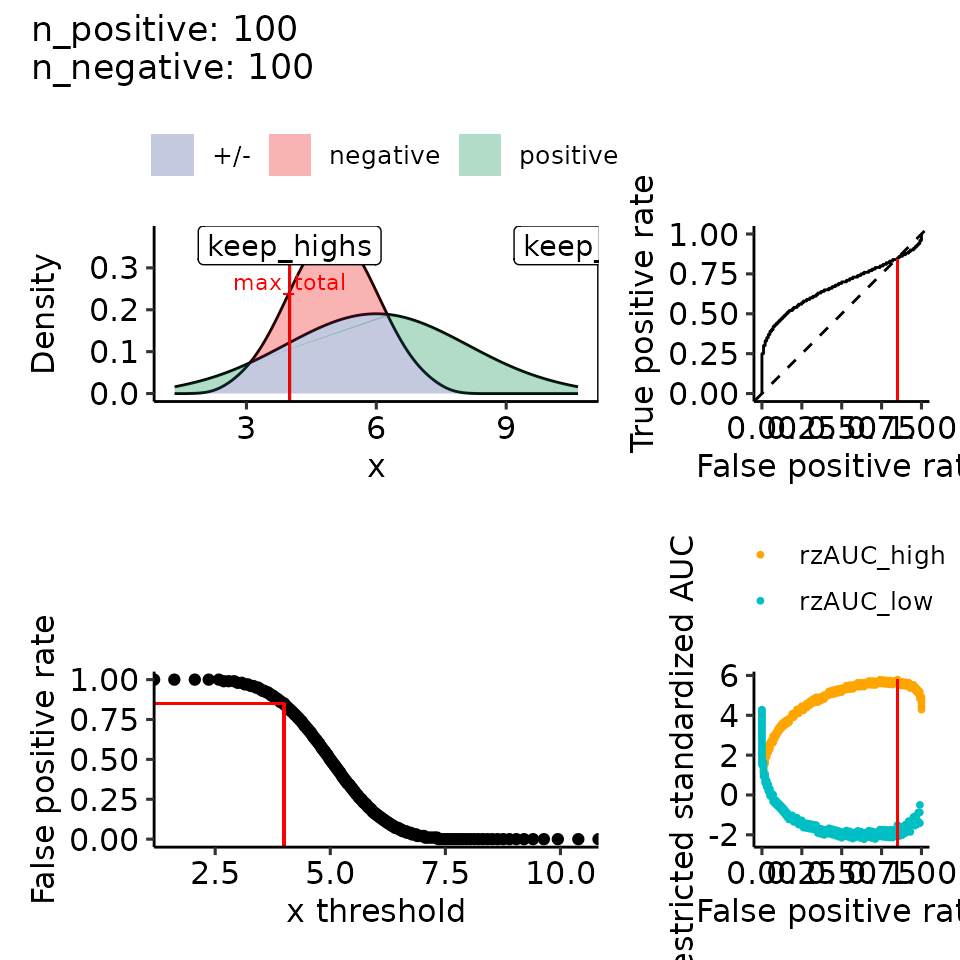
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""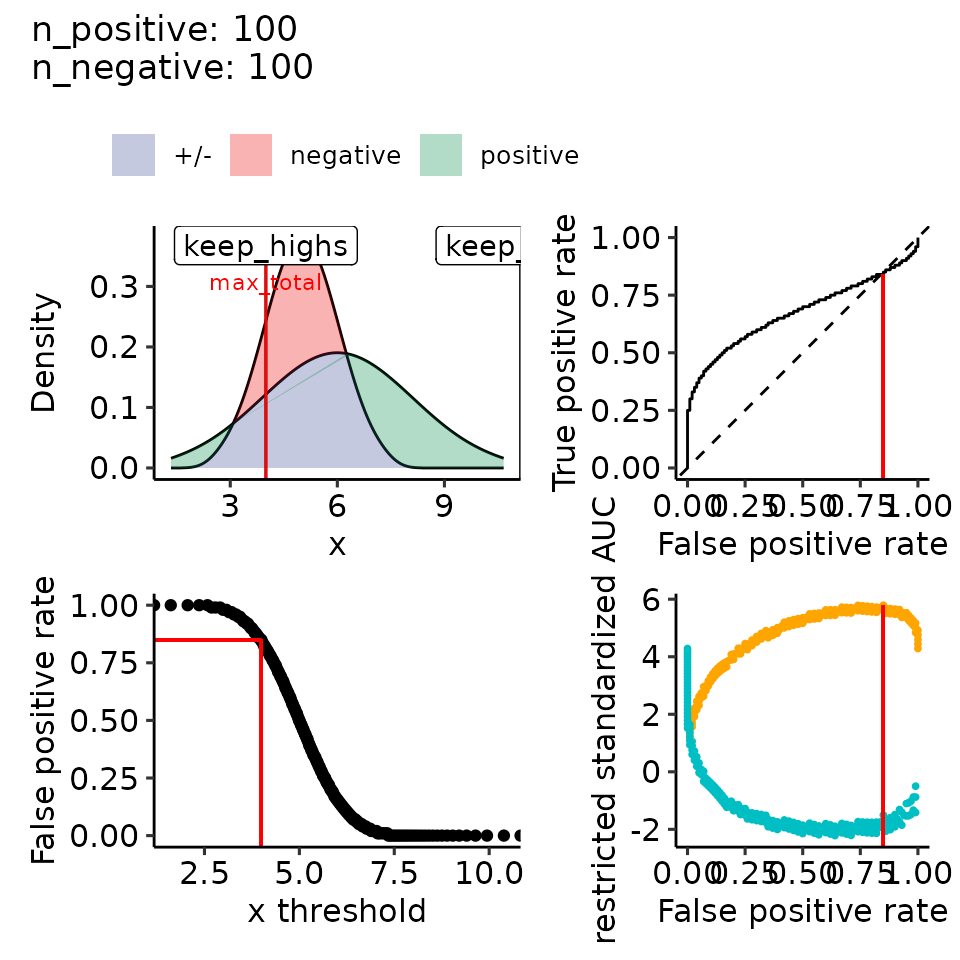
#>
#> [[3]]
#>
#> [[4]]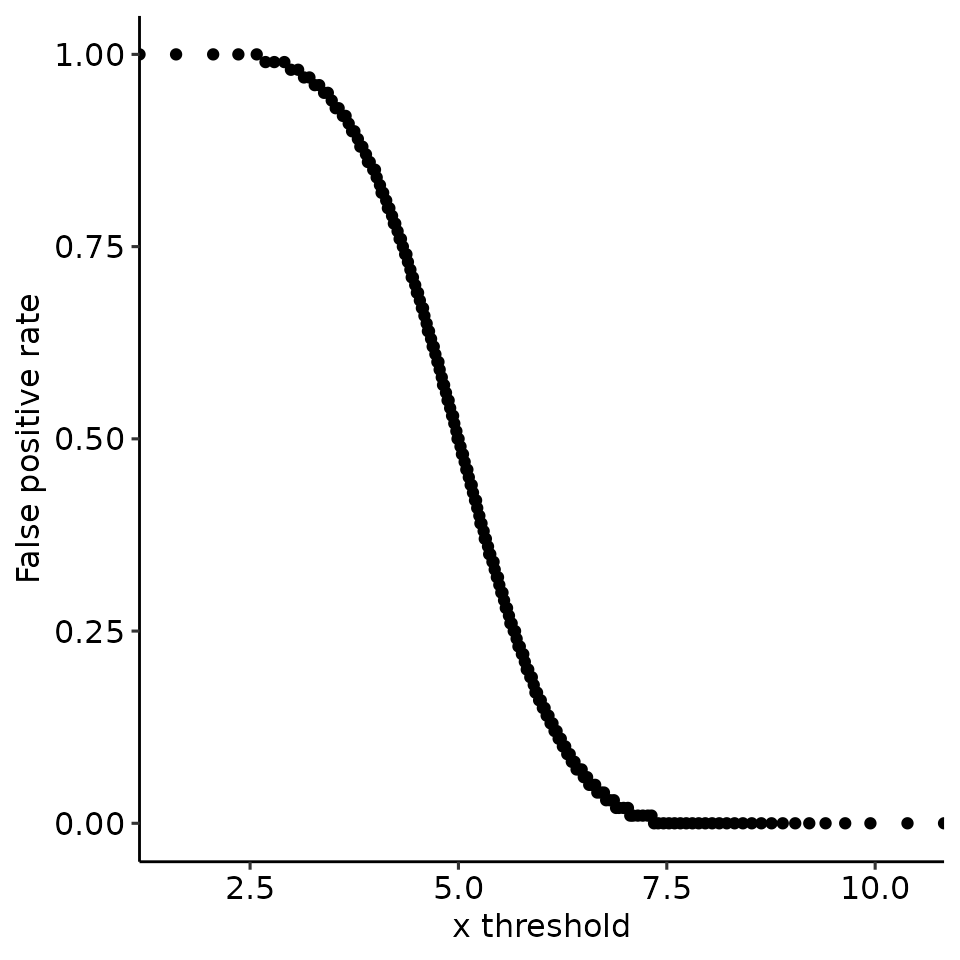
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""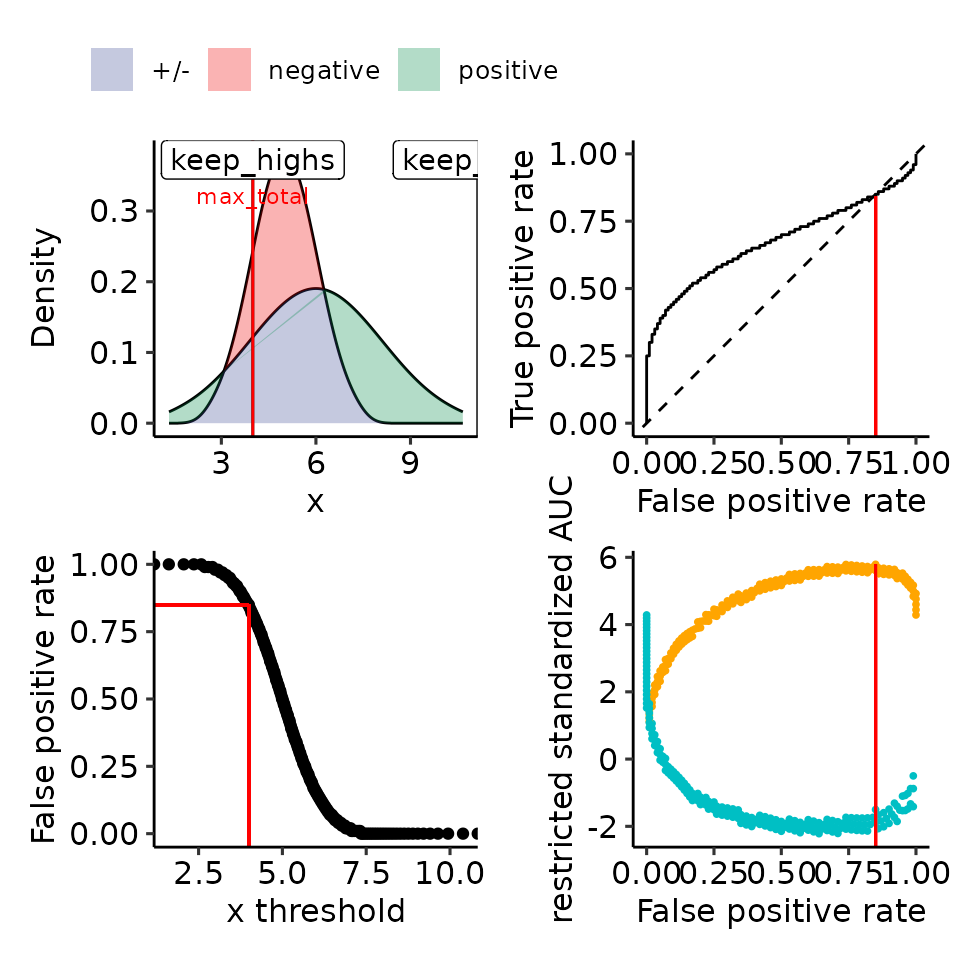
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 1.339842 1.884288 2.229646 2.489399 2.700655 2.880440 3.038055
#> [8] 3.179161 3.307475 3.425573 3.535318 3.638106 3.735007 3.826864
#> [15] 3.914352 3.998020 4.078324 4.155644 4.230300 4.302569 4.372686
#> [22] 4.440858 4.507265 4.572066 4.635401 4.697396 4.758164 4.817806
#> [29] 4.876415 4.934075 4.990861 5.046846 5.102093 5.156664 5.210614
#> [36] 5.263995 5.316857 5.369246 5.421205 5.472777 5.524000 5.574913
#> [43] 5.625552 5.675952 5.726148 5.776171 5.826054 5.875830 5.925528
#> [50] 5.975181 6.024819 6.074472 6.124170 6.173946 6.223829 6.273852
#> [57] 6.324048 6.374448 6.425087 6.476000 6.527223 6.578795 6.630754
#> [64] 6.683143 6.736005 6.789386 6.843336 6.897907 6.953154 7.009139
#> [71] 7.065925 7.123585 7.182194 7.241836 7.302604 7.364599 7.427934
#> [78] 7.492735 7.559142 7.627314 7.697431 7.769700 7.844356 7.921676
#> [85] 8.001980 8.085648 8.173136 8.264993 8.361894 8.464682 8.574427
#> [92] 8.692525 8.820839 8.961945 9.119560 9.299345 9.510601 9.770354
#> [99] 10.115712 10.660158
#>
#> $data$negative
#> [1] 2.669921 2.942144 3.114823 3.244699 3.350327 3.440220 3.519027 3.589580
#> [9] 3.653737 3.712786 3.767659 3.819053 3.867503 3.913432 3.957176 3.999010
#> [17] 4.039162 4.077822 4.115150 4.151284 4.186343 4.220429 4.253633 4.286033
#> [25] 4.317700 4.348698 4.379082 4.408903 4.438208 4.467037 4.495431 4.523423
#> [33] 4.551047 4.578332 4.605307 4.631997 4.658428 4.684623 4.710603 4.736388
#> [41] 4.762000 4.787457 4.812776 4.837976 4.863074 4.888085 4.913027 4.937915
#> [49] 4.962764 4.987591 5.012409 5.037236 5.062085 5.086973 5.111915 5.136926
#> [57] 5.162024 5.187224 5.212543 5.238000 5.263612 5.289397 5.315377 5.341572
#> [65] 5.368003 5.394693 5.421668 5.448953 5.476577 5.504569 5.532963 5.561792
#> [73] 5.591097 5.620918 5.651302 5.682300 5.713967 5.746367 5.779571 5.813657
#> [81] 5.848716 5.884850 5.922178 5.960838 6.000990 6.042824 6.086568 6.132497
#> [89] 6.180947 6.232341 6.287214 6.346263 6.410420 6.480973 6.559780 6.649673
#> [97] 6.755301 6.885177 7.057856 7.330079
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""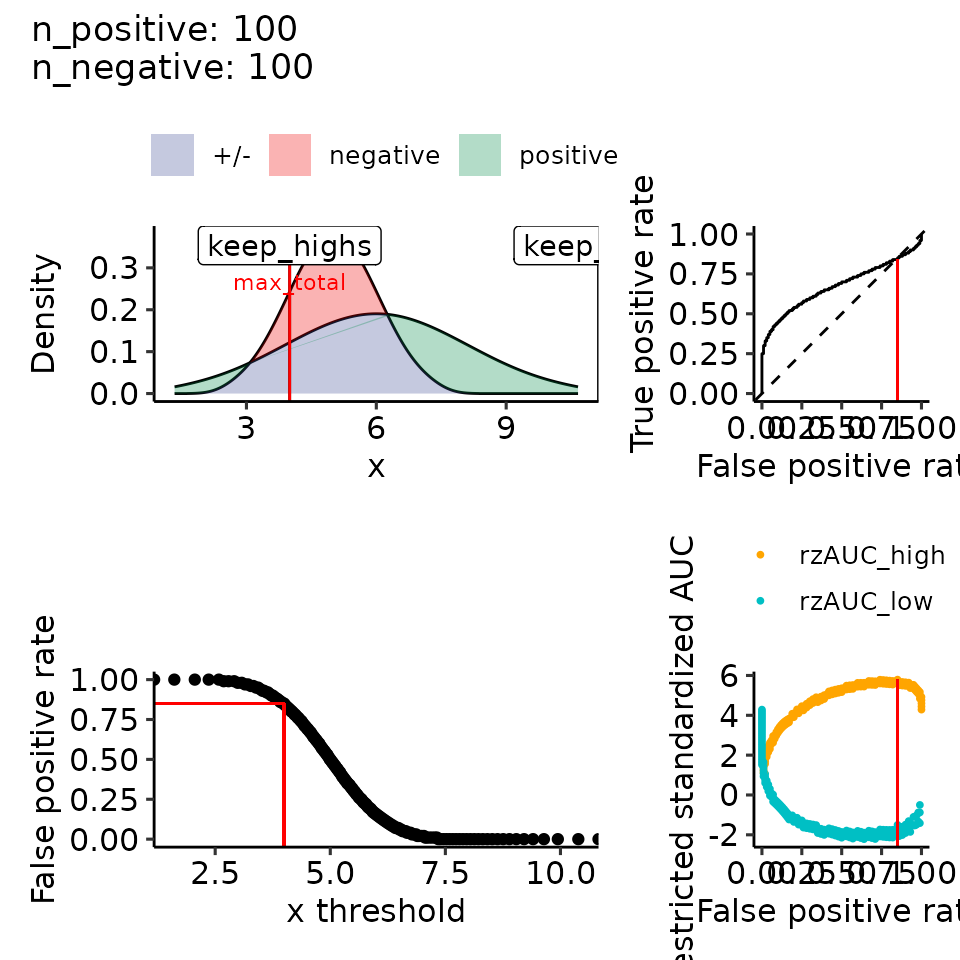
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 201 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.675 100 100 1 0.00168
#> 2 1.61 0.682 99 100 1.01 0.00168
#> 3 2.06 0.689 98 100 1.02 0.00169
#> 4 2.36 0.696 97 100 1.03 0.00170
#> 5 2.58 0.704 96 100 1.04 0.00171
#> 6 2.69 0.701 96 99 1.05 0.00172
#> 7 2.79 0.708 95 99 1.06 0.00173
#> 8 2.91 0.715 94 99 1.07 0.00174
#> 9 2.99 0.713 94 98 1.09 0.00175
#> 10 3.08 0.720 93 98 1.10 0.00176
#> # ℹ 191 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.6754 0.6754 4.285704 1.821613e-05
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.7579832 0.001984127 5.791708 6.967407e-09 3.998515
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.6754 0.001675 4.285704 1.821613e-05 Inf
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.7579832 0.001984127 5.791708 6.967407e-09 3.998515 high
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.6754
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()
# 4.2 mean: positive > negative, var: positive < negative --> right-skewed
filename <- file.path(paste0(main_plotname, "_posGTneg_posVARLTneg.pdf"))
# pdf(filename, height = main_height * size_factor, width = main_width * size_factor)
rroc_theo <- plot_rROC_theoretical(
qnorm_positive = function(x) qnorm(x, mean = 6, sd = 1),
qnorm_negative = function(x) qnorm(x, mean = 5, sd = 2),
n_positive = n_positives,
n_negative = n_negatives,
return_all = TRUE
)
print(plots_including_fpr(rroc_theo))
#> [[1]]
#> Ignoring unknown labels:
#> • colour : ""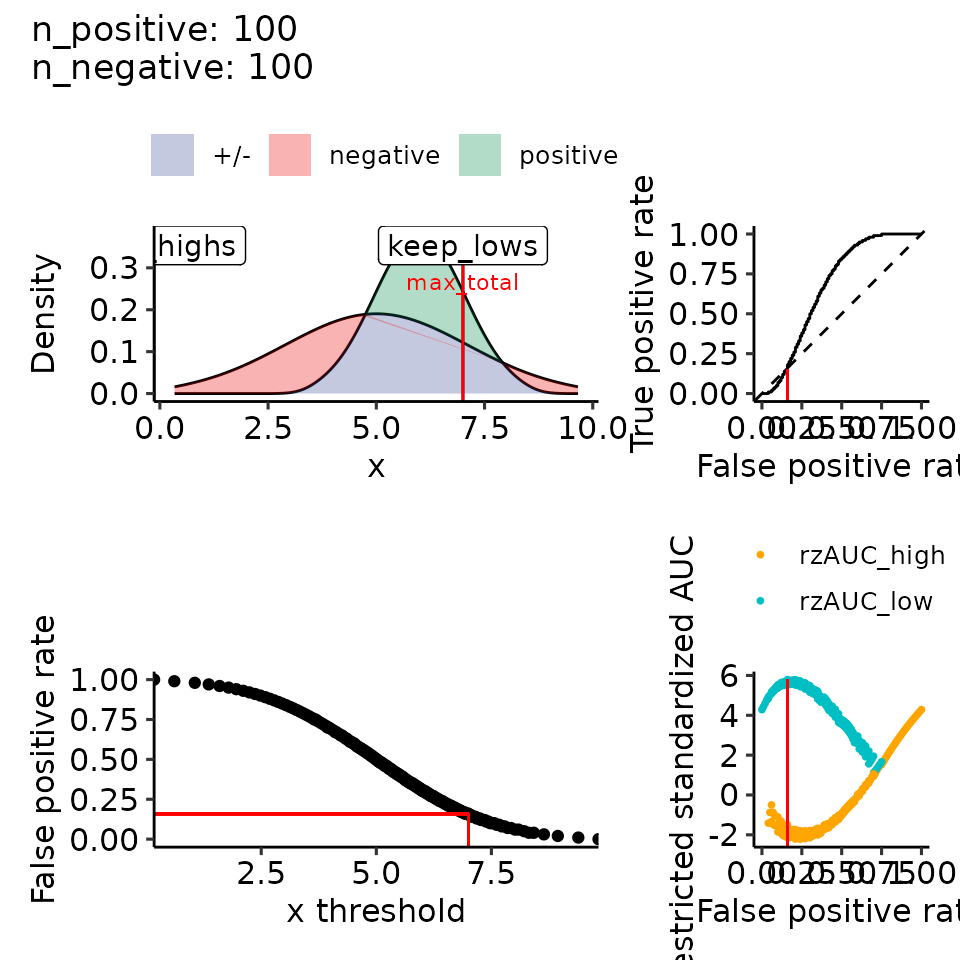
#>
#> [[2]]
#> Ignoring unknown labels:
#> • colour : ""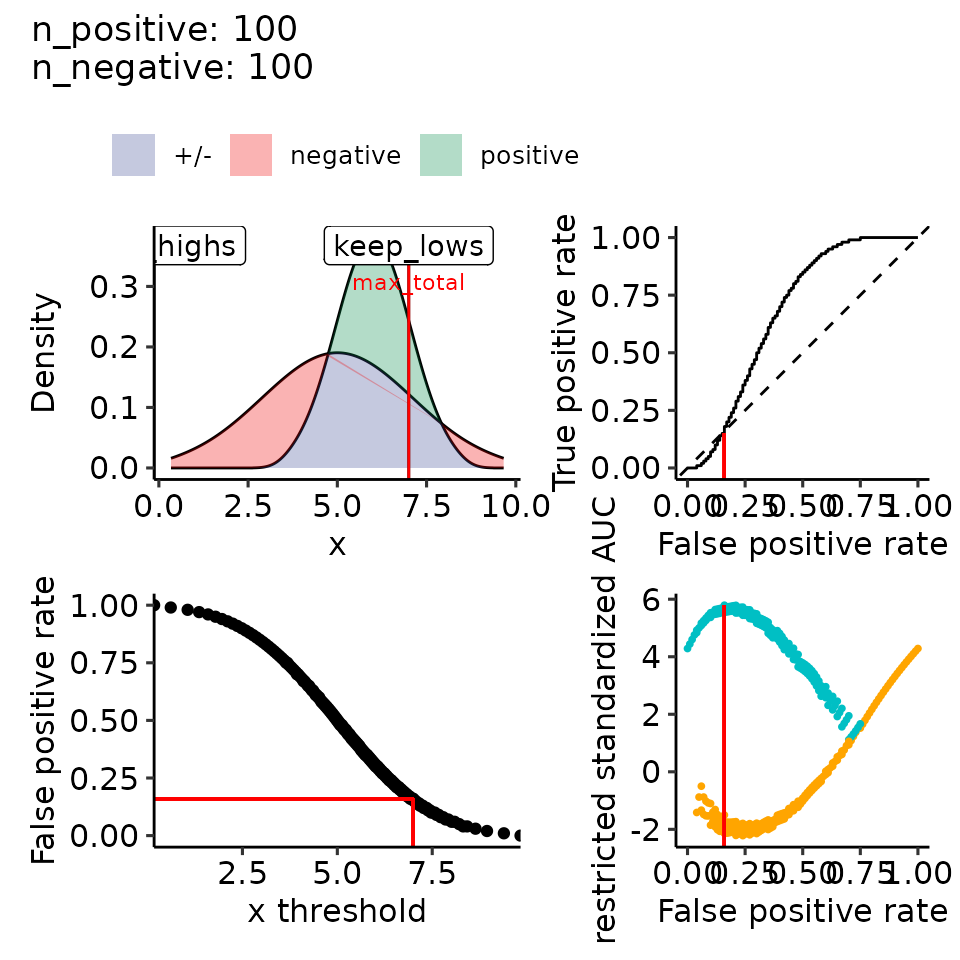
#>
#> [[3]]
#>
#> [[4]]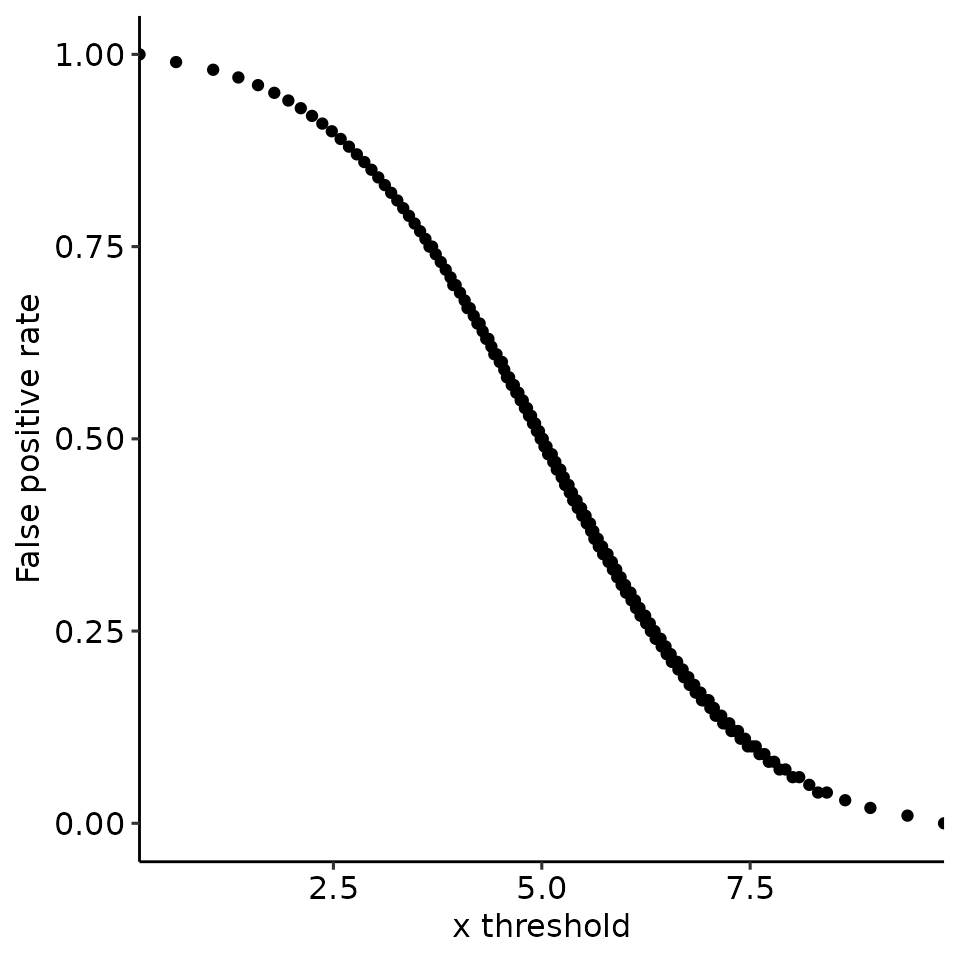
#>
#> [[5]]
#> Ignoring unknown labels:
#> • colour : ""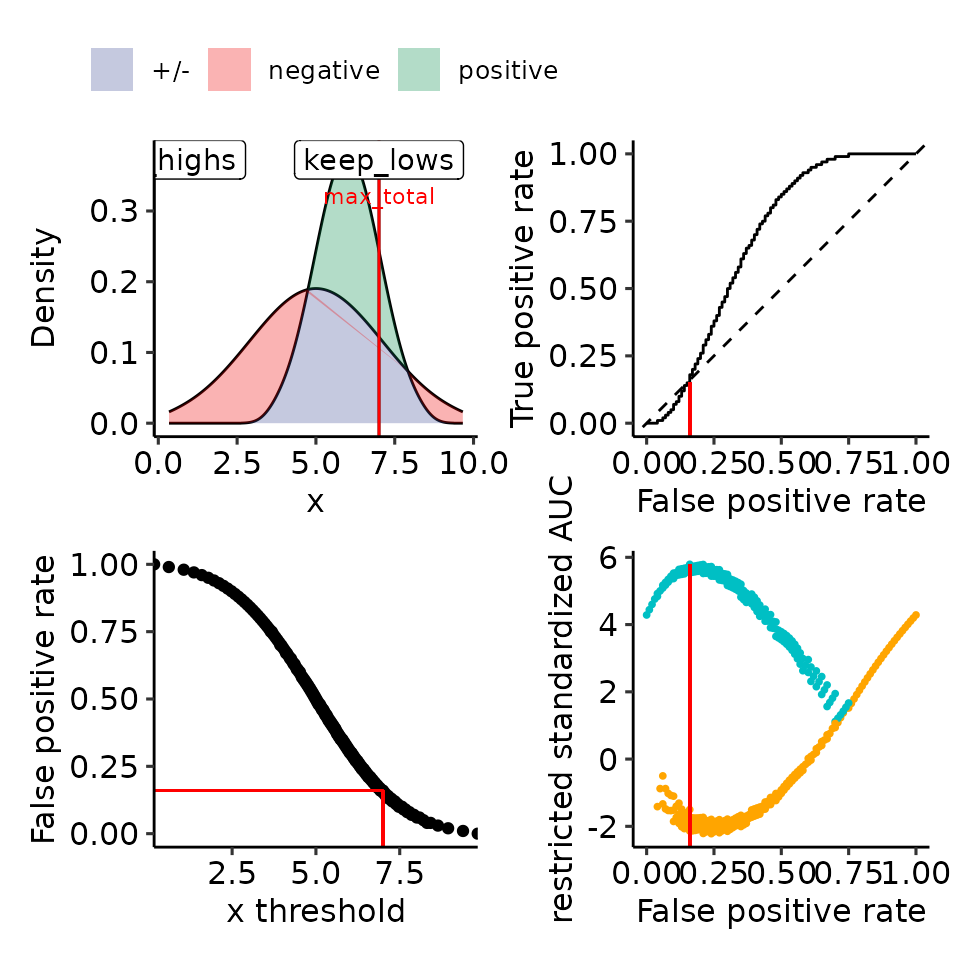
# dev.off()
# sink(paste0(filename, ".txt"))
print(rroc_theo)
#> $data
#> $data$positive
#> [1] 3.669921 3.942144 4.114823 4.244699 4.350327 4.440220 4.519027 4.589580
#> [9] 4.653737 4.712786 4.767659 4.819053 4.867503 4.913432 4.957176 4.999010
#> [17] 5.039162 5.077822 5.115150 5.151284 5.186343 5.220429 5.253633 5.286033
#> [25] 5.317700 5.348698 5.379082 5.408903 5.438208 5.467037 5.495431 5.523423
#> [33] 5.551047 5.578332 5.605307 5.631997 5.658428 5.684623 5.710603 5.736388
#> [41] 5.762000 5.787457 5.812776 5.837976 5.863074 5.888085 5.913027 5.937915
#> [49] 5.962764 5.987591 6.012409 6.037236 6.062085 6.086973 6.111915 6.136926
#> [57] 6.162024 6.187224 6.212543 6.238000 6.263612 6.289397 6.315377 6.341572
#> [65] 6.368003 6.394693 6.421668 6.448953 6.476577 6.504569 6.532963 6.561792
#> [73] 6.591097 6.620918 6.651302 6.682300 6.713967 6.746367 6.779571 6.813657
#> [81] 6.848716 6.884850 6.922178 6.960838 7.000990 7.042824 7.086568 7.132497
#> [89] 7.180947 7.232341 7.287214 7.346263 7.410420 7.480973 7.559780 7.649673
#> [97] 7.755301 7.885177 8.057856 8.330079
#>
#> $data$negative
#> [1] 0.3398422 0.8842880 1.2296459 1.4893990 1.7006546 1.8804400 2.0380547
#> [8] 2.1791609 2.3074747 2.4255725 2.5353183 2.6381059 2.7350069 2.8268638
#> [15] 2.9143515 2.9980202 3.0783241 3.1556436 3.2303003 3.3025689 3.3726864
#> [22] 3.4408585 3.5072653 3.5720658 3.6354007 3.6973958 3.7581637 3.8178060
#> [29] 3.8764151 3.9340746 3.9908614 4.0468460 4.1020934 4.1566639 4.2106136
#> [36] 4.2639948 4.3168567 4.3692455 4.4212051 4.4727768 4.5240002 4.5749133
#> [43] 4.6255524 4.6759524 4.7261475 4.7761707 4.8260542 4.8758298 4.9255285
#> [50] 4.9751813 5.0248187 5.0744715 5.1241702 5.1739458 5.2238293 5.2738525
#> [57] 5.3240476 5.3744476 5.4250867 5.4759998 5.5272232 5.5787949 5.6307545
#> [64] 5.6831433 5.7360052 5.7893864 5.8433361 5.8979066 5.9531540 6.0091386
#> [71] 6.0659254 6.1235849 6.1821940 6.2418363 6.3026042 6.3645993 6.4279342
#> [78] 6.4927347 6.5591415 6.6273136 6.6974311 6.7696997 6.8443564 6.9216759
#> [85] 7.0019798 7.0856485 7.1731362 7.2649931 7.3618941 7.4646817 7.5744275
#> [92] 7.6925253 7.8208391 7.9619453 8.1195600 8.2993454 8.5106010 8.7703541
#> [99] 9.1157120 9.6601578
#>
#>
#> $rroc
#> $rroc$plots
#> Ignoring unknown labels:
#> • colour : ""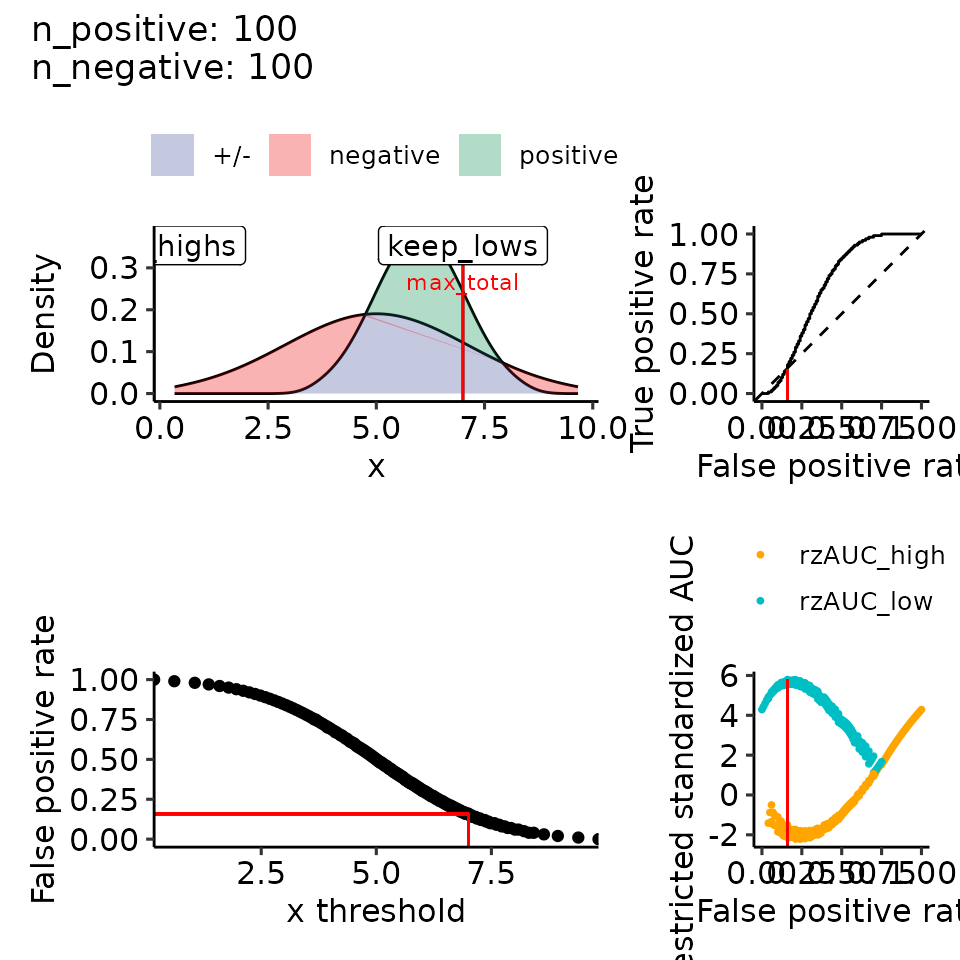
#>
#> $rroc$single_rROC
#> $performances
#> # A tibble: 201 × 21
#> threshold auc_high positives_high negatives_high scaling_high auc_var_H0_high
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 -Inf 0.675 100 100 1 0.00168
#> 2 0.612 0.672 100 99 1.01 0.00168
#> 3 1.06 0.669 100 98 1.02 0.00169
#> 4 1.36 0.665 100 97 1.03 0.00170
#> 5 1.60 0.662 100 96 1.04 0.00171
#> 6 1.79 0.658 100 95 1.05 0.00172
#> 7 1.96 0.655 100 94 1.06 0.00173
#> 8 2.11 0.651 100 93 1.08 0.00174
#> 9 2.24 0.647 100 92 1.09 0.00175
#> 10 2.37 0.643 100 91 1.10 0.00176
#> # ℹ 191 more rows
#> # ℹ 15 more variables: rzAUC_high <dbl>, pval_asym_onesided_high <dbl>,
#> # pval_asym_high <dbl>, auc_low <dbl>, positives_low <dbl>,
#> # negatives_low <dbl>, scaling_low <dbl>, auc_var_H0_low <dbl>,
#> # rzAUC_low <dbl>, pval_asym_onesided_low <dbl>, pval_asym_low <dbl>,
#> # tp <dbl>, fp <dbl>, tpr_global <dbl>, fpr_global <dbl>
#>
#> $global
#> auc auc_var_H0 rzAUC pval_asym
#> 1 0.6754 0.6754 4.285704 1.821613e-05
#>
#> $keep_highs
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.6754 0.001675 4.285704 1.821613e-05 -Inf
#>
#> $keep_lows
#> auc auc_var_H0 rzAUC pval_asym threshold
#> 1 0.7579832 0.001984127 5.791708 6.967407e-09 7.001485
#>
#> $max_total
#> auc auc_var_H0 rzAUC pval_asym threshold part
#> 1 0.7579832 0.001984127 5.791708 6.967407e-09 7.001485 low
#>
#> $positive_label
#> [1] "positive"
#>
#> $pROC_full
#>
#> Call:
#> roc.default(response = true_pred_df[["true"]], predictor = true_pred_df[["pred"]], levels = c(FALSE, TRUE), direction = direction)
#>
#> Data: true_pred_df[["pred"]] in 100 controls (true_pred_df[["true"]] FALSE) < 100 cases (true_pred_df[["true"]] TRUE).
#> Area under the curve: 0.6754
#>
#> attr(,"class")
#> [1] "restrictedROC" "list"
# sink()